WiscSIMS Importer
Design philosophy and context
The WiscSIMS laboratory is a National Facility for Stable Isotope Geochemistry, supported by NSF. The lab measures a wide variety of isotope systems on an equally wide range of samples. Some analyses are commonly performed (e.g. δ18O) but new analytical methods are developed commonly. Hence, the data import pipeline must be flexible enough to expand to new methods and changes to the data tables. In addition, in archival data, some metadata must be inferred from human entered text that is not uniform from user-to-user.
Necessary analytical data information from files
- Measured values
- Standard-Sample-Standard bracket structure
- Mount name
- Running standard
Creating uniform columns
Uniform columns are created using a dictionary of column names. Below is an example snippet of the table. The DictionaryColNames column is a list of exact matches for column names to be relabeled with the ColNames entry. A specific subset of column names is set with the SIMSmethods column. The unit and Type colums are used later on import. REQUIRED is still in development, but aims to allow for adding new columns without causing failures on import for files that do not contain those columns.
| ColNames | DictionaryColNames | SIMSmethods | unit | Type | REQUIRED |
|---|---|---|---|---|---|
| File | File; Filename | d18O10; d13C7 | Filename | Text | TRUE |
| Comment | Comment; Comment...2 | d18O10; d13C7 | Comment | Text | TRUE |
| d18OVSMOW | \u03B418O \u2030 VSMOW vs UWC-3; d18O \u2030 VSMOW; d18_VSMOW; d18O [VSMOW],d18 VSMOW; \u03B418O \u2030 VSMOW; 18O VSMOW vs UWC-3; δ18O ‰ VSMOW vs UWC-3; d18O ‰ VSMOW; δ18O ‰ VSMOW | d18O10 | permille_VSMOW | Numeric | FALSE |
| SD2ext | 2SD (ext.); Er (2S); 2SD; Std_1SD; Er(2S) | d18O10; d13C7 | permille | Numeric | FALSE |
Identifying analyses of standards
A large number of standards have been developed for the WiscSIMS lab because the method requires similar composition and structure between standard and sample. In the image below, the standards are identifiable by the UWC-3 designation, and the blanks for columns C through F.
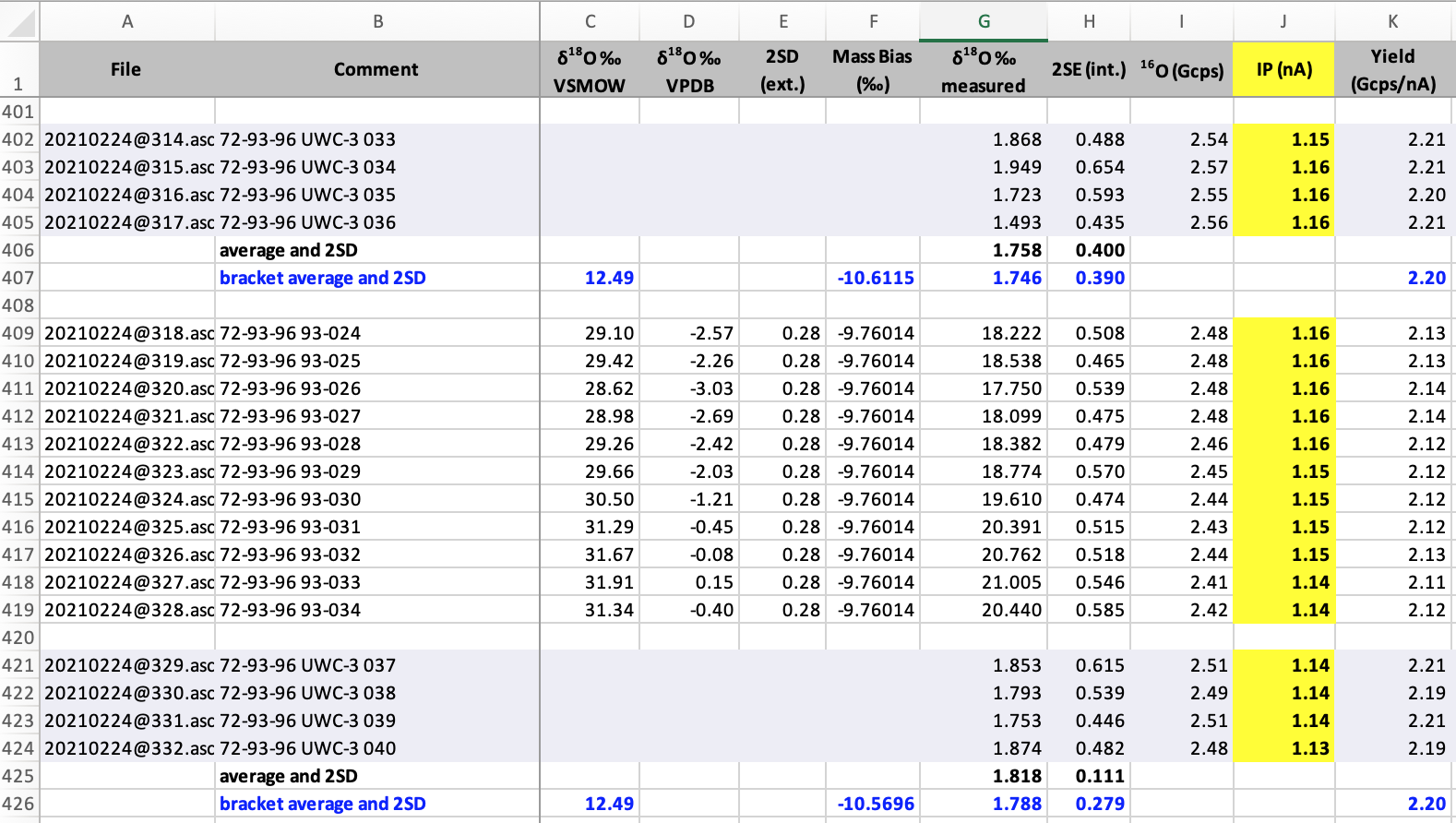
So in the importer, analyses of standards can be identified in several ways. The first is using regular expression. See code snippet below.
MaterialRegex <- "UW|-WI|KIM|SC"
Output$MATERIAL <-
ifelse(
grepl(MaterialRegex, Output$Comment, ignore.case = TRUE),
'STD',
ifelse(
!grepl(MaterialRegex, Output$Comment, ignore.case = TRUE),
'Sample',
NA
)
)
Standards in this example are identified by having the strings UW, -WI, KIM, _or _SC, in the comment column. This can be easily expanded and but should also be checked against other criteria, including a lack of standardized isotope ratio value and number of sequential analyses.
Standard-Sample-Standard brackets
Identification of brackets in the SIMS datasets are critical. This is done using the following procedure which looks for runs of the same entry (e.g. STD or Sample) and then notes the row where it changes. Doing this allows for re-calculation of standard corrected isotope ratios given identification of the running standard.
run_index <- with(
rle(material),
data.frame(
number = values,
start = cumsum(lengths) - lengths + 1,
end = cumsum(lengths)
)[order(values), ]
)
Standard regular expression
Identification of specific standards is done using regular expression. A table of standards including a name, calibrated isotope values, regular expression for lookup, standard designation (e.g. running/calibration), and material type (e.g. carbonate/silicate) is used for lookup. Regular expression for the UWC-3 standard is UWC\D*3 and each comment column in the original data sheet is checked for this string. Then matches have a column added with the standard name as identified by the regular expression (see code below for loop).
for (i in 1:nrow(Standards)) {
Output$RegexSTD[grepl(Standards$REGEX[i], Output$Comment, ignore.case = TRUE)] <-
Standards$StdName[i]
Output$is_standard[grepl(Standards$REGEX[i], Output$Comment, ignore.case = TRUE)] <-
TRUE
}
Connecting to Sparrow
Once data are cleaned and reformatted, they need to be handed-off to Sparrow. Because these operations have been done in R, the connection is slightly different than other labs.
Creating JSON in R
Creating JSON in R that fit the criteria necessary for Sparrow upload is slightly tedious. Data must be structured using nested lists to properly convert to JSON.
DatumList [[m]] <- list(
value = 1,
type = list(
parameter = "d18O_vsmow",
unit = "permille_vsmow"
)
)
The above code shows an example of this process. The DatumList contains 1 for value and then another list with the name type that contains both the name of the parameter and the unit that the parameter is measured in. The [[m]] notation is used after DatumList because it is in a for loop that works through the numeric values in the initial dataframe. We split initial excel files into multiple sessions because multiple SIMS mounts are typically used in a session and the Sparrow session model is a single instrument run on a sample.
The full reformatting code for the Session data model is three nested loops to populate nested lists. Full code for the SparrowReformatter.R function can be found on github.
Docker and R
Currently this code is executed in its own Docker container. To set this up a Dockerfile must be created. Our version is below. We use FROM r-base to load R in the container. It is loaded using Rocker. More documentation can be found on this website.
FROM r-base
LABEL version="1.1"
RUN R -e "install.packages(c('readxl', 'plyr'), dependencies=TRUE, repos='http://cran.rstudio.com')"
RUN R -e "install.packages(c('jsonlite'), dependencies=TRUE, repos='http://cran.rstudio.com')"
RUN R -e "install.packages(c('tidyr'), dependencies=TRUE, repos='http://cran.rstudio.com')"
RUN R -e "install.packages(c('purr'), dependencies=TRUE, repos='http://cran.rstudio.com')"
RUN apt-get update && apt-get install libssl-dev libcurl4-openssl-dev -y
RUN apt-get update && apt-get install -y libxml2-dev
RUN R -e "install.packages('httr')"
RUN R -e "install.packages(c('rlist','xml2'))"
RUN R -e "install.packages('stringr')"
RUN R -e "install.packages('dplyr')"
COPY *.R /code/
COPY **/*.R /code/
WORKDIR /code/
We need to install needed packages into the Docker container. This is done in the Docker container using the following code:RUN R -e "install.packages('package_name', dependencies=TRUE, repos='http://cran.rstudio.com')"In the larger code block above above, additional dependencies outside of R are necessary for some packages. These include libxml2 and libssl.
The PUT request and error checking
Data are passed to Sparrow using a PUT to the api v1 backend currently "http://backend:5000/api/v1/import-data/session" and the response is stored for error checking. Print out in the console is necessary with the print command.
response <-
PUT(url = "http://backend:5000/api/v1/import-data/session",
body = request,
encode = "json")
errors <- content(response)
print(errors)
Loading files in Docker container
We process multiple files at once with the importer. To do this, files must be visible to docker, and a script using the R data extraction pipeline are used. Files are made visible by changing the volumes: in the importer-container.yaml file. For instance in the development branch of the WiscSIMS project, the following is used to tie the OnlySIMSExcel folder on the desktop to a folder with the name AllTestData in the Docker container.
version: "3.6"
services:
importer:
#image: r-base
build: ../importer
volumes:
- "/Users/macrostrat/Desktop/OnlySIMSExcel:/AllTestData"
Looping through files
The short script executed by sparrow import-data is shown below. This script wraps together the scripts described above and creates a version of the DatumNesting function (SparrowReformater.R) that fails without ending the script. This is done using purr::possibly().
Some filtering for appropriate Excel files is also done in this script by finding $$\delta^{18}O$$or $$\delta^{13}C$$in its current form.
library(jsonlite)
library(httr)
library(tidyr)
library(readxl)
library(stringr)
library(purrr)
library(dplyr)
source("SparrowReformater.R")
possibly_DatumNesting <- possibly(DatumNesting, otherwise = "failed to load")
FileDirectory <- '/AllTestData/'
FileList <- as.vector(list.files(path = FileDirectory, pattern = ".xl?", recursive = TRUE))
ExcelFileData <- FileList %>% separate(FileList, c("Date", "Isotope", "User"), "_")
ExcelFileData <- cbind(FullPath, FileList, ExcelFileData)
ExcelFileDataCulled <- ExcelFileData[ExcelFileData$Isotope=='d18O'|ExcelFileData$Isotope=='d13C',]
ExcelFileDataCulled <- ExcelFileDataCulled[!is.na(ExcelFileDataCulled$Isotope),]
for(i in 1:nrow(ExcelFileDataCulled))
{
print(paste("Parse and upload ",
FileDirectory,
as.character(ExcelFileDataCulled$FullPath[i]), sep=""))
possibly_DatumNesting(paste(FileDirectory2,
as.character(ExcelFileDataCulled$FullPath[i]), sep=""))
}